Inital comit.
Basic transfer function and tank impedance plotting
This commit is contained in:
commit
190ca4ded5
12 changed files with 4221 additions and 0 deletions
92
pySmithPlot/.gitignore
vendored
Normal file
92
pySmithPlot/.gitignore
vendored
Normal file
|
|
@ -0,0 +1,92 @@
|
|||
# Byte-compiled / optimized / DLL files
|
||||
__pycache__/
|
||||
*.py[cod]
|
||||
*$py.class
|
||||
|
||||
# C extensions
|
||||
*.so
|
||||
|
||||
# Distribution / packaging
|
||||
.Python
|
||||
env/
|
||||
build/
|
||||
develop-eggs/
|
||||
dist/
|
||||
downloads/
|
||||
eggs/
|
||||
.eggs/
|
||||
lib/
|
||||
lib64/
|
||||
parts/
|
||||
sdist/
|
||||
var/
|
||||
*.egg-info/
|
||||
.installed.cfg
|
||||
*.egg
|
||||
|
||||
# PyInstaller
|
||||
# Usually these files are written by a python script from a template
|
||||
# before PyInstaller builds the exe, so as to inject date/other infos into it.
|
||||
*.manifest
|
||||
*.spec
|
||||
|
||||
# Installer logs
|
||||
pip-log.txt
|
||||
pip-delete-this-directory.txt
|
||||
|
||||
# Unit test / coverage reports
|
||||
htmlcov/
|
||||
.tox/
|
||||
.coverage
|
||||
.coverage.*
|
||||
.cache
|
||||
nosetests.xml
|
||||
coverage.xml
|
||||
*,cover
|
||||
.hypothesis/
|
||||
|
||||
# Translations
|
||||
*.mo
|
||||
*.pot
|
||||
|
||||
# Django stuff:
|
||||
*.log
|
||||
local_settings.py
|
||||
|
||||
# Flask stuff:
|
||||
instance/
|
||||
.webassets-cache
|
||||
|
||||
# Scrapy stuff:
|
||||
.scrapy
|
||||
|
||||
# Sphinx documentation
|
||||
docs/_build/
|
||||
|
||||
# PyBuilder
|
||||
target/
|
||||
|
||||
# IPython Notebook
|
||||
.ipynb_checkpoints
|
||||
|
||||
# pyenv
|
||||
.python-version
|
||||
|
||||
# celery beat schedule file
|
||||
celerybeat-schedule
|
||||
|
||||
# dotenv
|
||||
.env
|
||||
|
||||
# virtualenv
|
||||
venv/
|
||||
ENV/
|
||||
|
||||
# Spyder project settings
|
||||
.spyderproject
|
||||
|
||||
# Rope project settings
|
||||
.ropeproject
|
||||
|
||||
# Pycharm project settings
|
||||
.idea
|
||||
73
pySmithPlot/README.md
Executable file
73
pySmithPlot/README.md
Executable file
|
|
@ -0,0 +1,73 @@
|
|||
pySmithPlot
|
||||
===========
|
||||
|
||||
## New Release of Version 0.2
|
||||
|
||||
After 2 years of getting dusty **pySmithPlot** now got some new features and bug fixes. Here is a short changelog:
|
||||
|
||||
- **Support for Python 3**
|
||||
- improved grid generation algorithm
|
||||
- plot() now also handles also single numbers and purely real data
|
||||
- plot() can now interpolate lines between points or generate an equidistant spacing
|
||||
- changed handling of input data and renormalization; now the actual datatype (S,Z,Y-Parameter) can be specified when calling plot()
|
||||
- changed behaviour for normalization and placement of the label
|
||||
- added some parameter checks
|
||||
- removed default `matplotlib` settings
|
||||
- renamed some parameters to improve consistency
|
||||
- fixed issues with Unicode symbols
|
||||
- fixed issues with grid generation
|
||||
- fixed issues with axis label display and placement
|
||||
|
||||
There are still some plans for the future and they hopefully don't take another two years:
|
||||
|
||||
- [ ] support for Admittance Charts
|
||||
- [ ] support for `contour()` plots
|
||||
- [ ] zoom and 'cut out' function
|
||||
- [ ] special handling of other `matplotlib.patch` objects like arrows
|
||||
- [ ] ...
|
||||
|
||||
## Features
|
||||
|
||||
**pySmithPlot** is a matplotlib extension providing a projection class for creating high quality Smith Charts with Python. The generated plots blend seamless into matplotlib's style and support almost the full range of customization options.
|
||||
|
||||
This Library allows the fully automatic generation of Smith Charts with various customizable parameters and well selected default values. It also provides the following modifications and extensions:
|
||||
|
||||
- circle shaped drawing area with labels placed around
|
||||
- plot() accepts real and complex numbers as well as numpy.ndarray's
|
||||
- lines can be automatically interpolated to improve the optical appearance
|
||||
- data ranges can be interpolated to an equidistant spacing
|
||||
- start/end markers of lines can be modified and rotate tangential
|
||||
- gridlines are 3-point arcs to improve space efficiency of exported plots
|
||||
- 'fancy' option for adaptive grid generation
|
||||
- own tick locators for nice axis labels
|
||||
|
||||
For making a Smith Chart plot, it is sufficient to `import smithplot` and create a new subplot with projection set to 'smith'. (Requires matplotlib version 1.2)
|
||||
|
||||
A short example can be found in the `testbenches` directory and started with:
|
||||
|
||||
python3 smith_short_test.py
|
||||
|
||||
For more details and documentation, take a look into `smithplot/smithaxes.py`.
|
||||
|
||||
`testbenches/smith_full_test.py` runs various testbenches and gives a comparison for almost all parameters. These are the generated sample plots:
|
||||
|
||||

|
||||
[Grid Styles - PDF](https://github.com/vMeijin/pySmithPlot/wiki/images/examples/sample_grid_styles.pdf)
|
||||
|
||||
|
||||
[Fancy Threshold - PDF](https://github.com/vMeijin/pySmithPlot/wiki/images/examples/sample_fancy_grid.pdf)
|
||||
|
||||

|
||||
[Grid Locators - PDF](https://github.com/vMeijin/pySmithPlot/wiki/images/examples/sample_grid_locators.pdf)
|
||||
|
||||

|
||||
[Marker Modification - PDF](https://github.com/vMeijin/pySmithPlot/wiki/images/examples/sample_markers.pdf)
|
||||
|
||||

|
||||
[Interpolation - PDF](https://github.com/vMeijin/pySmithPlot/wiki/images/examples/sample_interpolation.pdf)
|
||||
|
||||

|
||||
[Normalize - PDF](https://github.com/vMeijin/pySmithPlot/wiki/images/examples/sample_normalize.pdf)
|
||||
|
||||

|
||||
[Miscellaneous - PDF](https://github.com/vMeijin/pySmithPlot/wiki/images/examples/sample_miscellaneous.pdf)
|
||||
19
pySmithPlot/setup.py
Executable file
19
pySmithPlot/setup.py
Executable file
|
|
@ -0,0 +1,19 @@
|
|||
import os
|
||||
|
||||
from setuptools import setup
|
||||
|
||||
|
||||
def read(fname):
|
||||
return open(os.path.join(os.path.dirname(__file__), fname)).read()
|
||||
|
||||
|
||||
setup(name="pysmithplot",
|
||||
version="0.2.0",
|
||||
packages=["smithplot"],
|
||||
description="An extension for Matplotlib providing a projection class to generate high quality Smith Chart plots.",
|
||||
long_description=read('README.md'),
|
||||
author="Paul Staerke",
|
||||
author_email="paul.staerke@gmail.com",
|
||||
license="BSD",
|
||||
url="https://github.com/vMeijin/pySmithPlot",
|
||||
install_requires=["matplotlib >= 1.2.0", "numpy", "scipy"])
|
||||
12
pySmithPlot/smithplot/__init__.py
Normal file
12
pySmithPlot/smithplot/__init__.py
Normal file
|
|
@ -0,0 +1,12 @@
|
|||
# -*- coding: utf-8 -*-
|
||||
import matplotlib
|
||||
from matplotlib.projections import register_projection
|
||||
|
||||
from .smithaxes import SmithAxes
|
||||
|
||||
# check version requierment
|
||||
if matplotlib.__version__ < '1.2':
|
||||
raise ImportError("pySmithPlot requires at least matplotlib version 1.2")
|
||||
|
||||
# add smith projection to available projections
|
||||
register_projection(SmithAxes)
|
||||
1544
pySmithPlot/smithplot/smithaxes.py
Normal file
1544
pySmithPlot/smithplot/smithaxes.py
Normal file
File diff suppressed because it is too large
Load diff
60
pySmithPlot/smithplot/smithhelper.py
Normal file
60
pySmithPlot/smithplot/smithhelper.py
Normal file
|
|
@ -0,0 +1,60 @@
|
|||
# -*- coding: utf-8 -*-
|
||||
# last edit: 11.04.2018
|
||||
|
||||
from collections import Iterable
|
||||
|
||||
import numpy as np
|
||||
|
||||
INF = 1e9
|
||||
EPSILON = 1e-7
|
||||
TWO_PI = 2 * np.pi
|
||||
|
||||
|
||||
def xy_to_z(*xy):
|
||||
if len(xy) == 1:
|
||||
z = xy[0]
|
||||
if isinstance(z, Iterable):
|
||||
z = np.array(z)
|
||||
if len(z.shape) == 2:
|
||||
z = z[0] + 1j * z[1]
|
||||
elif len(z.shape) > 2:
|
||||
raise ValueError("Something went wrong!")
|
||||
elif len(xy) == 2:
|
||||
x, y = xy
|
||||
if isinstance(x, Iterable):
|
||||
if isinstance(y, Iterable) and len(x) == len(y):
|
||||
z = np.array(x) + 1j * np.array(y)
|
||||
else:
|
||||
raise ValueError("x and y vectors dont match in type and/or size")
|
||||
else:
|
||||
z = x + 1j * y
|
||||
else:
|
||||
raise ValueError("Arguments are not valid - specify either complex number/vector z or real and imaginary number/vector x, y")
|
||||
|
||||
return z
|
||||
|
||||
|
||||
def z_to_xy(z):
|
||||
return z.real, z.imag
|
||||
|
||||
|
||||
def moebius_z(*args, norm):
|
||||
z = xy_to_z(*args)
|
||||
return 1 - 2 * norm / (z + norm)
|
||||
|
||||
|
||||
def moebius_inv_z(*args, norm):
|
||||
z = xy_to_z(*args)
|
||||
return norm * (1 + z) / (1 - z)
|
||||
|
||||
|
||||
def ang_to_c(ang, radius=1):
|
||||
return radius * (np.cos(ang) + np.sin(ang) * 1j)
|
||||
|
||||
|
||||
def lambda_to_rad(lmb):
|
||||
return lmb * 4 * np.pi
|
||||
|
||||
|
||||
def rad_to_lambda(rad):
|
||||
return rad * 0.25 / np.pi
|
||||
2
pySmithPlot/testbenches/.gitignore
vendored
Normal file
2
pySmithPlot/testbenches/.gitignore
vendored
Normal file
|
|
@ -0,0 +1,2 @@
|
|||
export.pdf
|
||||
|
||||
1002
pySmithPlot/testbenches/data/s11.csv
Executable file
1002
pySmithPlot/testbenches/data/s11.csv
Executable file
File diff suppressed because it is too large
Load diff
1002
pySmithPlot/testbenches/data/s22.csv
Executable file
1002
pySmithPlot/testbenches/data/s22.csv
Executable file
File diff suppressed because it is too large
Load diff
265
pySmithPlot/testbenches/smith_full_test.py
Executable file
265
pySmithPlot/testbenches/smith_full_test.py
Executable file
|
|
@ -0,0 +1,265 @@
|
|||
#!/usr/bin/env python3
|
||||
|
||||
import os
|
||||
import shutil
|
||||
import sys
|
||||
import time
|
||||
from multiprocessing.pool import Pool
|
||||
from types import FunctionType
|
||||
|
||||
import numpy as np
|
||||
from matplotlib import rcParams, pyplot as pp
|
||||
|
||||
sys.path.append("..")
|
||||
from smithplot.smithaxes import SmithAxes
|
||||
from smithplot import smithhelper
|
||||
|
||||
rcParams.update({"legend.numpoints": 3,
|
||||
"axes.axisbelow": True})
|
||||
|
||||
# sample data
|
||||
steps = 40
|
||||
data = np.loadtxt("data/s11.csv", delimiter=",", skiprows=1)[::steps]
|
||||
sp_data = data[:, 1] + data[:, 2] * 1j
|
||||
|
||||
data = np.loadtxt("data/s22.csv", delimiter=",", skiprows=1)[::steps]
|
||||
z_data = 50 * (data[:, 1] + data[:, 2] * 1j)
|
||||
|
||||
# default params
|
||||
SmithAxes.update_scParams({"plot.marker.hack": False,
|
||||
"plot.marker.rotate": False,
|
||||
"grid.minor.enable": False,
|
||||
"grid.minor.fancy": False})
|
||||
|
||||
FT = [False, True]
|
||||
figsize = 6
|
||||
ExportFormats = ["pdf", "png"]
|
||||
|
||||
|
||||
def plot_example(testbench, title, scale=50, **kwargs):
|
||||
print("Testbench '%s' : %s" % (testbench, title.replace("\n", "")))
|
||||
kwargs.setdefault("markevery", 1)
|
||||
pp.plot(smithhelper.moebius_inv_z(sp_data, norm=50), datatype="Z", **kwargs)
|
||||
pp.plot(z_data, datatype="Z", **kwargs)
|
||||
pp.plot(100, datatype="Z", **kwargs)
|
||||
pp.plot(25 + 25j, datatype="Z", **kwargs)
|
||||
pp.title(title)
|
||||
|
||||
|
||||
def savefig(testbench):
|
||||
for ext in ExportFormats:
|
||||
pp.savefig("%s/sample_%s.%s" % (build_path, testbench.lower().replace(" ", "_"), ext), format=ext)
|
||||
|
||||
|
||||
def tb_grid_styles():
|
||||
tb = "Grid Styles"
|
||||
fig = pp.figure(figsize=(3 * figsize, 2 * figsize))
|
||||
fig.set_tight_layout(True)
|
||||
|
||||
i = 0
|
||||
for major_fancy in FT:
|
||||
for minor in FT:
|
||||
for minor_fancy in FT:
|
||||
if minor or not minor_fancy:
|
||||
i += 1
|
||||
pp.subplot(2, 3, i, projection="smith",
|
||||
grid_major_fancy=major_fancy,
|
||||
grid_minor_enable=minor,
|
||||
grid_minor_fancy=minor_fancy)
|
||||
|
||||
major_str = "fancy" if major_fancy else "standard"
|
||||
minor_str = "off" if not minor else "fancy" if minor_fancy else "standard"
|
||||
|
||||
plot_example(tb, "Major: %s - Minor: %s" % (major_str, minor_str))
|
||||
|
||||
savefig(tb)
|
||||
|
||||
|
||||
def tb_fancy_grids():
|
||||
tb = "Fancy Grid"
|
||||
fig = pp.figure(figsize=(3 * figsize, 2 * figsize))
|
||||
fig.set_tight_layout(True)
|
||||
|
||||
i = 0
|
||||
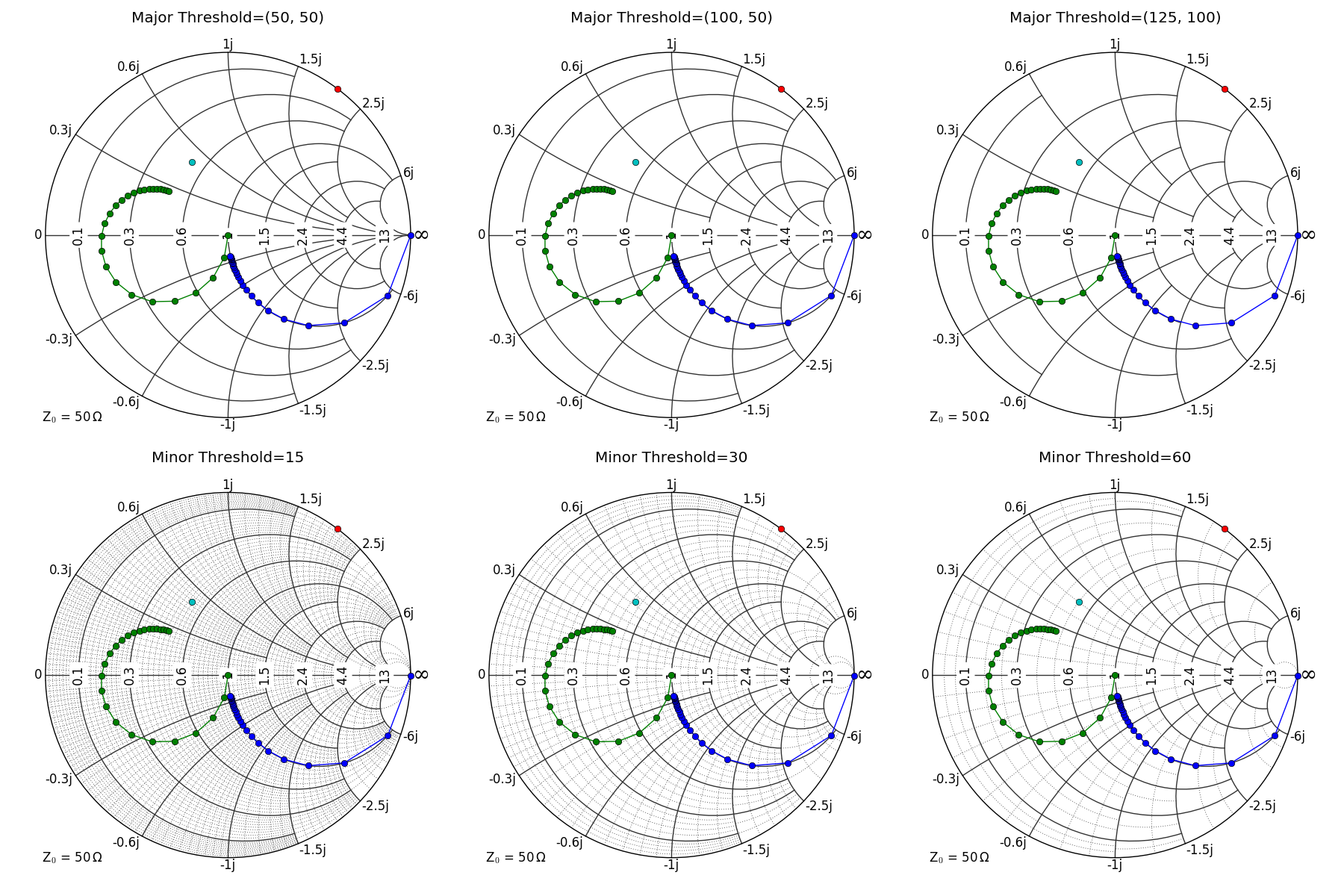
for threshold in [(50, 50), (100, 50), (125, 100)]:
|
||||
i += 1
|
||||
pp.subplot(2, 3, i, projection="smith",
|
||||
grid_major_fancy_threshold=threshold)
|
||||
plot_example(tb, "Major Threshold=(%d, %d)" % threshold)
|
||||
|
||||
for threshold in [15, 30, 60]:
|
||||
i += 1
|
||||
pp.subplot(2, 3, i, projection="smith",
|
||||
grid_minor_fancy=True,
|
||||
grid_minor_enable=True,
|
||||
grid_minor_fancy_threshold=threshold)
|
||||
plot_example(tb, "Minor Threshold=%d" % threshold)
|
||||
|
||||
savefig(tb)
|
||||
|
||||
|
||||
def tb_grid_locators():
|
||||
tb = "Grid Locators"
|
||||
fig = pp.figure(figsize=(4 * figsize, 2 * figsize))
|
||||
fig.set_tight_layout(True)
|
||||
|
||||
i = 0
|
||||
for num in [5, 8, 14, 20]:
|
||||
i += 1
|
||||
pp.subplot(2, 4, i, projection="smith",
|
||||
grid_major_xmaxn=num)
|
||||
plot_example(tb, "Max real steps: %d" % num)
|
||||
|
||||
for num in [6, 14, 25, 50]:
|
||||
i += 1
|
||||
pp.subplot(2, 4, i, projection="smith",
|
||||
grid_major_ymaxn=num)
|
||||
plot_example(tb, "Max imaginary steps: %d" % num)
|
||||
|
||||
savefig(tb)
|
||||
|
||||
|
||||
def tb_normalize():
|
||||
tb = "Normalize"
|
||||
fig = pp.figure(figsize=(3 * figsize, 2 * figsize))
|
||||
fig.set_tight_layout(True)
|
||||
|
||||
i = 0
|
||||
for normalize in FT:
|
||||
for impedance in [10, 50, 200]:
|
||||
i += 1
|
||||
pp.subplot(2, 3, i, projection="smith",
|
||||
axes_impedance=impedance,
|
||||
axes_normalize=normalize)
|
||||
plot_example(tb, "Impedance: %d Ω — Normalize: %s" % (impedance, normalize))
|
||||
|
||||
savefig(tb)
|
||||
|
||||
|
||||
def tb_markers():
|
||||
tb = "Markers"
|
||||
VStartMarker = np.array([[0, 0], [0.5, 0.5], [0, -0.5], [-0.5, 0.5], [0, 0]])
|
||||
XEndMarker = np.array([[0, 0], [0.5, 0.5], [0.25, 0], [0.5, -0.5], [0, 0], [-0.5, -0.5], [-0.25, 0], [-0.5, 0.5], [0, 0]])
|
||||
|
||||
fig = pp.figure(figsize=(4 * figsize, 2 * figsize))
|
||||
fig.set_tight_layout(True)
|
||||
|
||||
i = 0
|
||||
for hackline, startmarker, endmarker, rotate_marker in [[False, None, None, False],
|
||||
[True, "s", "^", False],
|
||||
[True, "s", None, False],
|
||||
[True, VStartMarker, XEndMarker, False],
|
||||
[True, "s", "^", True],
|
||||
[True, None, "^", False]]:
|
||||
i += 1
|
||||
ax = pp.subplot(2, 3, i, projection="smith",
|
||||
plot_marker_hack=hackline,
|
||||
plot_marker_rotate=rotate_marker)
|
||||
SmithAxes.update_scParams(instance=ax, plot_marker_start=startmarker,
|
||||
plot_marker_end=endmarker)
|
||||
|
||||
def ptype(x):
|
||||
if isinstance(x, np.ndarray):
|
||||
return "custom"
|
||||
elif x is True:
|
||||
return "on"
|
||||
elif x is False:
|
||||
return "off"
|
||||
elif x is None:
|
||||
return None
|
||||
else:
|
||||
return "'%s'" % x
|
||||
|
||||
plot_example(tb, "HackLines: %s - StartMarker: %s\nEndMarker: %s - Rotate: %s" % tuple(map(ptype, [hackline, startmarker, endmarker, rotate_marker])), markersize=10)
|
||||
|
||||
savefig(tb)
|
||||
|
||||
|
||||
def tb_interpolation():
|
||||
tb = "Interpolation"
|
||||
fig = pp.figure(figsize=(3 * figsize, 2 * figsize))
|
||||
fig.set_tight_layout(True)
|
||||
|
||||
i = 0
|
||||
for interpolation, equipoints in [[False, False],
|
||||
[10, False],
|
||||
[False, 10],
|
||||
[False, 50]]:
|
||||
i += 1
|
||||
pp.subplot(2, 2, i, projection="smith")
|
||||
plot_example(tb, "Interpolation: %s — Equipoints: %s" % ("False" if interpolation is False else interpolation,
|
||||
"False" if equipoints is False else equipoints), interpolate=interpolation, equipoints=equipoints)
|
||||
|
||||
savefig(tb)
|
||||
|
||||
|
||||
def tb_misc():
|
||||
tb = "Miscellaneous"
|
||||
fig = pp.figure(figsize=(3 * figsize, 2 * figsize))
|
||||
fig.set_tight_layout(True)
|
||||
|
||||
pp.subplot(2, 3, 1, projection="smith",
|
||||
plot_marker_hack=True)
|
||||
plot_example(tb, "Legend")
|
||||
pp.legend(["S11", "S22", "Polyline", "Z \u2192 0.125l/\u03BB"])
|
||||
|
||||
divs = [1, 3, 7]
|
||||
pp.subplot(2, 3, 2, projection="smith",
|
||||
grid_minor_enable=True,
|
||||
grid_minor_fancy=True,
|
||||
grid_minor_fancy_dividers=divs)
|
||||
plot_example(tb, "Minor fancy dividers=%s" % divs)
|
||||
|
||||
pp.subplot(2, 3, 3, projection="smith",
|
||||
axes_radius=0.3)
|
||||
plot_example(tb, "Axes radius: 0.25")
|
||||
|
||||
pp.subplot(2, 3, 4, projection="smith",
|
||||
symbol_infinity="Inf",
|
||||
symbol_infinity_correction=0,
|
||||
symbol_ohm="Ohm")
|
||||
plot_example(tb, "Infinity symbol: 'Inf' — Ohm symbol: Ohm")
|
||||
|
||||
pp.subplot(2, 3, 5, projection="smith",
|
||||
grid_locator_precision=4)
|
||||
plot_example(tb, "Grid Locator Precision: 4")
|
||||
|
||||
pp.subplot(2, 3, 6, projection="smith",
|
||||
axes_xlabel_rotation=0)
|
||||
plot_example(tb, "Axes X Label Rotation: 0")
|
||||
|
||||
savefig(tb)
|
||||
|
||||
|
||||
build_all = True
|
||||
build_path = "./build"
|
||||
|
||||
if __name__ == '__main__':
|
||||
# clear and create path
|
||||
if os.path.exists(build_path):
|
||||
shutil.rmtree(build_path)
|
||||
time.sleep(0.5)
|
||||
os.makedirs(build_path)
|
||||
|
||||
if build_all:
|
||||
print("Start parallel testbenches...")
|
||||
p = Pool()
|
||||
r = []
|
||||
for key, func in locals().copy().items():
|
||||
if isinstance(func, FunctionType) and "tb_" in key:
|
||||
r += [p.apply_async(func, {})]
|
||||
|
||||
for proc in r:
|
||||
proc.get()
|
||||
else:
|
||||
pass
|
||||
# tb_grid_styles()
|
||||
# tb_fancy_grids()
|
||||
# tb_grid_locators()
|
||||
# tb_normalize()
|
||||
tb_markers()
|
||||
# tb_interpolation()
|
||||
# tb_misc()
|
||||
pp.show()
|
||||
|
||||
print("build finished")
|
||||
37
pySmithPlot/testbenches/smith_short_test.py
Executable file
37
pySmithPlot/testbenches/smith_short_test.py
Executable file
|
|
@ -0,0 +1,37 @@
|
|||
#!/usr/bin/env python3
|
||||
|
||||
import sys
|
||||
|
||||
import numpy as np
|
||||
from matplotlib import rcParams, pyplot as pp
|
||||
|
||||
rcParams.update({"legend.numpoints": 3})
|
||||
|
||||
sys.path.append("..")
|
||||
from smithplot import SmithAxes
|
||||
|
||||
# sample data
|
||||
data = np.loadtxt("data/s11.csv", delimiter=",", skiprows=1)[::100]
|
||||
val1 = data[:, 1] + data[:, 2] * 1j
|
||||
|
||||
data = np.loadtxt("data/s22.csv", delimiter=",", skiprows=1)[::100]
|
||||
val2 = data[:, 1] + data[:, 2] * 1j
|
||||
|
||||
# plot data
|
||||
pp.figure(figsize=(6, 6))
|
||||
|
||||
ax = pp.subplot(1, 1, 1, projection='smith')
|
||||
pp.plot([10, 100], markevery=1)
|
||||
|
||||
|
||||
pp.plot(200 + 100j, datatype=SmithAxes.Z_PARAMETER)
|
||||
pp.plot(50 * val1, label="default", datatype=SmithAxes.Z_PARAMETER)
|
||||
pp.plot(50 * val2, markevery=1, label="interpolate=3", interpolate=3, datatype=SmithAxes.Z_PARAMETER)
|
||||
pp.plot(val1, markevery=1, label="equipoints=22", equipoints=22, datatype=SmithAxes.S_PARAMETER)
|
||||
pp.plot(val2, markevery=3, label="equipoints=22, \nmarkevery=3", equipoints=22, datatype=SmithAxes.S_PARAMETER)
|
||||
|
||||
leg = pp.legend(loc="lower right", fontsize=12)
|
||||
pp.title("Matplotlib Smith Chart Projection")
|
||||
|
||||
pp.savefig("export.pdf", format="pdf", bbox_inches="tight")
|
||||
pp.show()
|
||||
Loading…
Add table
Add a link
Reference in a new issue